1. Introduction to Oce
Dan Kelley (https://orcid.org/0000-0001-7808-5911) and Clark Richards (https://orcid.org/0000-0002-7833-206X)
2026-02-17
Source:vignettes/A_oce.Rmd
A_oce.RmdAbstract. The oce package makes it easy
to read, summarize and plot data from a variety of Oceanographic
instruments, isolating the researcher from the quirky data formats that
are common in this field. It also provides functions for working with
basic seawater properties such as the equation of state, and with
derived quantities such as the buoyancy frequency. Although simple
enough to be used in a teaching context, oce is powerful
enough for a research setting. These things are illustrated here in a
general context, with further detail being provided by other vignettes,
by the documentation for the package’s functions and datasets, in Kelley
(2018) and in Kelley et al. (2022).
Introduction
Oceanographers must deal with measurements made by a wide variety of instruments, a task that is complicated by a tendency of instrument manufacturers to invent new data formats to suite new measurement capabilities. Many manufacturers provide software for scanning data files and producing overview plots, but it is of somewhat limited use to researchers who work with several instrument types at the same time, and who need to move beyond standardized engineering plots to specialized scientific plots and statistical analyses.
Architecture of oce objects
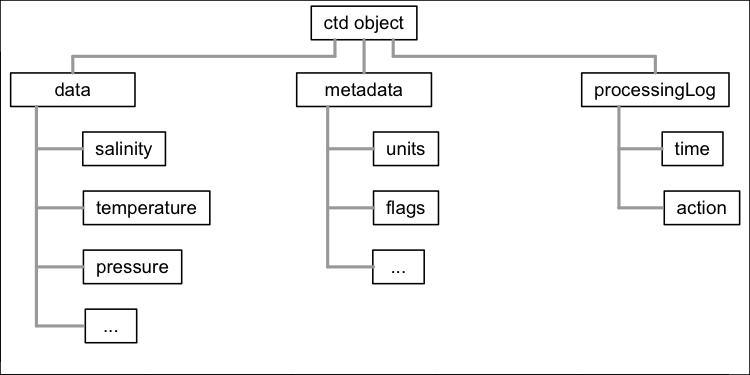
oce objects contain slots named
data, metadata, and
processingLog, with the contents depending on the type of
data.The need to scan diverse data files was one motivation for the
creation of oce, but an equal goal was to make it easy to
work with the data once they are in the system. This was accomplished
partly by the provision of specialized and generic (overloaded)
functions to work with the data, and partly by providing accessor
methods that make it convenient to reach inside the data objects (see
next section).
As illustrated in Figure 1, each oce object contains
three slots:
-
data, a list containing the actual data, e.g., for a CTD object (see also the vignette about CTD data), this will containpressure,temperature, etc. -
metadata, a list containing information about the data, such as units, data-quality flags, sampling locations, etc. -
processingLog, a list that documents how the object was created and possibly changed thereafter.
Accessing data in oce objects
For detailed analysis, users may want to access data within
oce objects. While it is possible to descend through the object using
the “slot” and “list” notation (e.g. d@data$salinity yields
the salinity within an oce object named d), this approach
is not recommended, for two reasons. First, direct access makes a user’s
code brittle to changes in the object structure (which are sometimes
necessary to track changes in measurement technologies, or in analysis
methods). And, second, direct access is limited to what is actually
stored in the file, and this might not be what the user wants – for
example, a record might contain electrical conductivity but not
salinity.
The preferred approach is to use the [[ notation, which
derives from the generic “Extract” method for accessing parts of an
object. It permits accessing not just the data stored in the object, but
also quantities that can be computed from such data (e.g. salinity, if
temperature, pressure and conductivity are stored).
A general introduction is provided by
?`[[,oce-method`and details are provided for individual object classes with e.g.
?`[[,ctd-method`for the ctd class.
The notation is simple. For example, suppose that d is
an object that stores salinity in either its
data slot or metadata slot. Then,
S <- d[["salinity"]]will extract the salinity if it is present in the object, or it will compute salinity, if the object’s contents are sufficient to compute salinity.
The [[ method first looks in the metadata
slot, but if the item is not found there, it proceeds to look in the
data slot. This two-step scheme is helpful because it frees
the user from having to know where data are stored. For example, in a
ctd object, latitude might be stored in the
metadata (for a CTD profile) or in the data
slot (for the slantwise sampling of a glider).
The [[ notation is helpful for several reasons, among
which two are prominent.
It can give valuable performance enhancements in some cases, e.g. the data in the
landsatclass are stored in two byte-level arrays, which yields a marked improvement over the 8-byte arrays that R generally uses on a 64-bit machine. The[[assembles these byte-scale chunks into a conventional R numerical matrix, which can be quite convenient for users who wish to operate on the data without learning how to assemble the bytes.It provides a uniformity of notation that can be helpful to users. In
oce, objects derived from data files tend to hold just the information in those files, not derived information. For example, For example, CTD datasets often provide in-situ temperature but not potential temperature (since the latter is not measured). The in-situ temperature is found with e.g.d[["temperature"]], and so it seems natural to writed[["theta"]]to get the potential temperature. If this is done, then thectdversion of[[first looks in the data slot, and returnsthetaif it is found there, as may sometimes be the case (depending on the choice of the person who created the dataset). However, if it is not found, then[[callsswTheta()to calculate the value, and returns the result.
Finally, it is also possible to extract the entirety of either the
metadata or data slot, e.g.
data <- d[["data"]]yields the full data slot, which is a list with elements that can be
accessed in the conventional way, e.g. for a ctd object,
and
data$temperatureretrieves the temperature.
In addition to the above, [[ has a special entry that
can reveal not just the data in the object, but also the data that can
be computed from the object. It returns a list with items named
metadata, metadataDerived, data
and dataDerived. The Derived items are
specific not just to the object class, but also to the data stored
within the particular object. For example,
reveals that the square of the buoyancy frequency can be retrieved
with ctd[["N2"]], but if say the temperature were removed,
then "N2" would not have been listed.
Exercise 1. Show that removing temperature from the
built-in "ctd" dataset makes [["?"]] indicate
that N2 is no longer available.
Modifying data in oce objects
There are several schemes for modifying the data within
oce objects. For example, and by analogy with the
[[ notation of the previous section, the following
will store the excess over freezing temperature into the
ctd object.
Further information on this notation is provided by
?"[[<-,oce-method"The above works only within the data slot. To store
within the metadata slot, consider using e.g.
ctd[["metadata"]]$scientist <- "Dalhousie Oceanography 4120/5120 Class of 2003"sets the “scientist”.
For archival work, it is important to store reasons for changing
things in an object. Two functions are provided for this purpose:
oceSetData and oceSetMetadata. For example, a
better way to change the scientist might be to write
ctd <- oceSetMetadata(ctd,
name = "scientist",
value = "Dalhousie Oceanography 4120/5120 Class of 2003",
note = "give credit where it's due"
)and a better way to store temperatureAboveFreezing would
be
ctd <- oceSetData(ctd,
name = "temperatureAboveFreezing",
value = ctd[["temperature"]] - swTFreeze(ctd),
unit = list(unit = expression(degree * C), scale = "ITS-90"),
originalName = "-",
note = "add temperatureAboveFreezing, for ice-related calculations"
)which illustrates that this notation, as opposed to the
[[<- notation, permits the specification of a unit and
an originalName, both of which, together with the
note, are displayed by summary(ctd).
Oce functions
The uniformity of the various oce objects helps users
build skill in examining and modifying objects. Fine-scale control is
provided throughout oce, but the best way to learn is to
start with simplest tools and their defaults. For example, the following
will read a CTD file named "station1.cnv", summarize the
contents, and plot an overview of the data, with profiles, a TS diagram,
and a map (Figure 2).
The reader should stop now and try this on a file of their own. The
pattern will work with a fairly wide variety of file types, because
read.oce() examines the file name and contents to try to
discover what it is. For an example, if read.oce() is given
the name of a file created by an Acoustic Doppler Profiler, it will
return an object inheriting from class "adp", so the
summary() and plot() calls will be tailored to
that type, e.g. the graph will show images of time-distance variation in
each of the measured velocity components.
Notes on oce function names.
As just illustrated, the general function to read a dataset ends in
.oce, and the name is a signal that the returned object is of classoce. Depending on the file contents,dwill also have an additional class, e.g. if the file contains CTD data, then the object would inherit from two classes,oceandctd, with the second being used to tailor the graphics by passing control to thectdvariant of the genericplotfunction (usehelp("plot,ctd-method")to learn more).Generally,
ocefunctions employ a “camel case” naming convention, in which a function that is described by several words is named by stringing the words together, capitalizing the first letter of second and subsequent words. For example,ctdFindProfiles()locates individual profiles within actdobject that holds data acquired during a cyclic raising and lowering of the CTD instrument.Function names begin with
ocein cases where a more natural name would be in conflict with a function in the base R system or a package commonly used by Oceanographers. For example,oceContour()is a function that provides an alternative tocontour()in thegraphicspackage.
Calculation of seawater properties
The oce package provides many functions for dealing with
seawater properties. Perhaps the most used is swRho(S,T,p),
which computes seawater density
,
where
is salinity,
is in-situ temperature in
C
(on the ITS-90 scale), and
is seawater pressure, i.e. the excess over atmospheric pressure, in
dbar. (This and similar functions starts with the letters
sw to designate that they relate to seawater properties.)
The result is a number in the order of
kg/m.
For many purposes, Oceanographers prefer to use the density anomaly
kg/m,
provided with swSigma(salinity,temperature,pressure), or
its adiabatic cousin
,
provided with swSigmaTheta().
Most of the functions can use either the UNESCO or GSW (TEOS-10)
formulation of seawater properties, with the choice set by an argument
called eos. It should be noted that oce uses
the gsw package for GSW calculations.
Caution: the results obtained with
eos="gsw" in oce functions may differ from the
results obtained when using the gsw functions directly, due
to unit conventions. For example, swCSTp(..., eos="gsw")
reports conductivity ratio for consistency with the UNESCO formulation,
however the underlying gsw function
gsw::gsw_C_from_SP() reports conductivity in mS/cm.
A partial list of seawater functions is as follows:
-
swAlpha()for thermal expansion coefficient -
swAlphaOverBeta()for -
swBeta()for haline compression coefficient -
swConductivity()for conductivity from , and -
swDepth()for depth from and latitude -
swDynamicHeight()for dynamic height -
swLapseRate()for adiabatic lapse rate -
swN2()for buoyancy frequency -
swRho()for density from , and -
swSCTp()for salinity from conductivity, temperature and pressure -
swSTrho()for salinity from temperature and density -
swSigma()for ,kg/m -
swSigmaT()for with set to zero and temperature unaltered -
swSigmaTheta()for with set to zero and temperature altered adiabatically -
swSoundSpeed()for speed of sound -
swSpecificHeat()for specific heat -
swSpice(),spiciness0(),spiciness1()andspiciness2()for quantities used in watermass research -
swTFreeze()for freezing temperature -
swTSrho()for temperature from salinity and density -
swTheta()for potential temperature -
swViscosity()for viscosity
Details and examples are provided in the documentation of these functions.
The following exercise may be of help to readers who prefer to learn by doing. (Answers are provided at the end of this document.)
Exercise 2. a. What is the density of a seawater parcel at pressure 100 dbar, with salinity 34 PSU and temperature 10C? b. What temperature would the parcel have if raised adiabatically to the surface? c. What density would it have if raised adiabatically to the surface? d. What density would it have if lowered about 100m, increasing the pressure to 200 dbar? e. Draw a blank - diagram with from 30 to 40 PSU and from -2 to 20C.
CTD data
The read.oce function recognizes a wide variety of CTD
data formats, and the associated plot function can produce
many types of graphical display. In addition, there are several
functions that aid in the analysis of such data. See the ctd vignette for more on dealing with CTD
data.
Section plots
The commands
will plot a summary diagram containing sections of
,
,
and
,
along with a chart indicating station locations. In addition to such
overview diagrams, the section variant of the generic
plot function can also create individual plots of
individual properties (use help("plot,section-method") to
learn more).
Some section datasets are supplied in a pre-gridded format, but it is
also common to have different pressure levels at each station. For such
cases, sectionGrid() may be used, e.g. the following
produces Figure 4. The ship was travelling westward from the
Mediterranean towards North America, taking 124 stations in total; the
stationId value selects the last few stations of the
section, during which the ship headed toward the northwest, crossing
isobaths (and perhaps, the Gulf Stream) at nearly right angles.
library(oce)
#> Loading required package: gsw
data(section)
GS <- subset(section, 102 <= stationId & stationId <= 124)
GSg <- sectionGrid(GS, p = seq(0, 1600, 25))
plot(GSg, which = c(1, 99), map.xlim = c(-85, -(64 + 13 / 60)))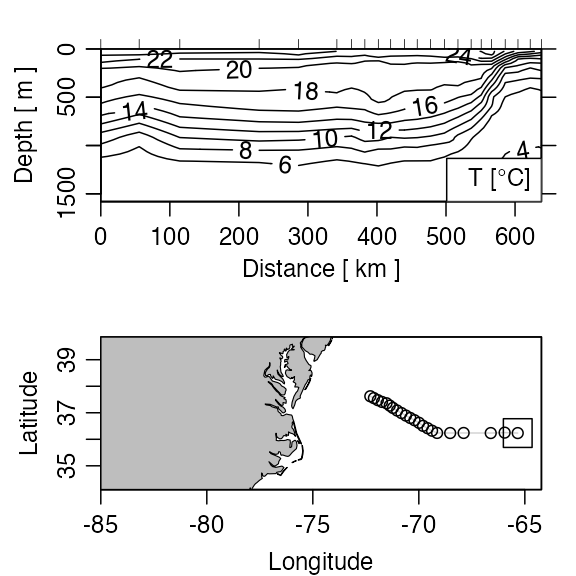
Figure 4. Portion of the CTD section designated A03, showing the Gulf Sream region. The square on the cruise track corresponds to zero distance on the section.
Exercise 3. Draw a
-
diagram for the section data, using black symbols to the east of 30W and
gray symbols to the west, thus highlighting Mediterranean-derived
waters. Use handleFlags() (see using
data-quality flags) to discard questionable data, and use the
accessor function [[.
Exercise 4. Plot dynamic height and geostrophic
velocity across the Gulf Stream. (Hint: use the
swDynamicHeight() function.)
Maps
Oce has several functions that facilitate the drawing of maps. A
variety of projections are provided, with the mathematics of projection
being handled behind the scenes with the sf package. An
introduction to drawing maps is provided with ?mapPlot, and
the map projection vignette
provides much more detail.
Acoustic Doppler Profiler data
The following commands produce Figure 10, showing one velocity
component of currents measured in the St Lawrence Estuary Internal Wave
Experiment. This plot type is just one of many provided by the
adp variant of the generic plot function (see
?"plot,adp-method"). See the adp
vignette for much more on acoustic Doppler profiler data.
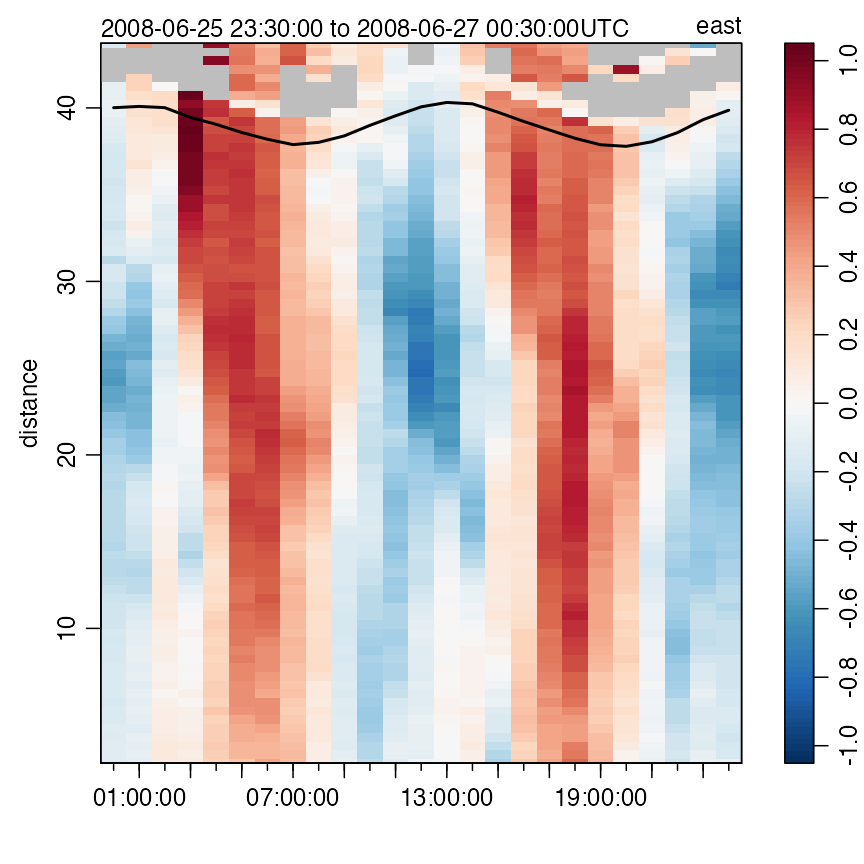
Figure 10. Measurements made with a bottom-mounted ADP in the St Lawrence Estuary. The line near the surface indicates pressure measured by the ADP.
Handling data-quality flags
Archives of CTD/bottle and Argo drifter measurements commonly supply data-quality flags that provide an indication of the trust to be put in individual data points. Oce has a flexible scheme for dealing with such flags, and also for inserting flags into these or any data type; see the using data-quality flags vignette for more..
Working in non-English languages
Many of the oce plotting functions produce axis labels
that can be displayed in languages other than English. At the time of
writing, French, German, Spanish, and Mandarin are supported in at least
a rudimentary way. Setting the language can be done at the general
system level, or within R, as indicated below (results not shown).
library(oce)
Sys.setenv(LANGUAGE = "fr")
data(ctd)
plot(ctd)Most of the translated items were found by online dictionaries, and so they may be incorrect for oceanographic usage. Readers can help out in the translation effort, if they have knowledge of how nautical words such as Pitch and Roll and technical terms such as Absolute Salinity and Potential Temperature should be written in various languages.
Extending oce
The oce object structure can be used as a basis for new object types.
This has the advantage the basic operations of oce will carry over to
the new types. For example, the accessing operator [[ and
summary function will work as expected, and so will such
aspects as the handling of data-quality flags and units. More details on
setting up classes that inherit from oce are provided in the subclassing vignette.
The future of oce
The present version of oce can only handle data types
that the authors have been using in their research. New data types will
be added as the need arises in that work, but the authors would also be
happy to add other data types that are likely to prove useful to the
Oceanographic community. (The data types need not be restricted to
Physical Oceanography, but the authors will need some help in dealing
with other types of data, given their research focus.)
Two principles will guide the addition of data types and functions: (a) the need, as perceived by the authors or by other contributors and (b) the ease with which the additions can be made. One might call this development-by-triage, by analogy to the scheme used in Emergency Rooms to organize medical effort efficiently.
Development website
The site https://github.com/dankelley/oce provides a
window on the development that goes on between the CRAN releases of the
package. Readers are requested to visit the site to report bugs, to
suggest new features, or just to see how oce development is
coming along. Note that the development branch is used by
the authors in their work, and is updated so frequently that it must be
considered unstable, at least in those spots being changed on a given
day. Official CRAN releases derive from the master branch,
and are done when the code is considered to be of reasonable stability
and quality. This is all in a common pattern for open-source
software.
Solutions to exercises
Exercise 1. Show that removing temperature from the
built-in "ctd" dataset makes [["?"]] indicate
that N2 is no longer available.
This may be demonstrated as below.
library(oce)
data(ctd)
"N2" %in% ctd[["?"]]$dataDerived
#> [1] TRUE
ctd@data$temperature <- NULL # erase temperature
"N2" %in% ctd[["?"]]$dataDerived
#> [1] FALSEExercise 2. Seawater properties. In the UNESCO system we may write
library(oce)
swRho(34, 10, 100, eos = "unesco")
#> [1] 1026.624
swTheta(34, 10, 100, eos = "unesco")
#> [1] 9.988599
swRho(34, swTheta(34, 10, 100, eos = "unesco"), 0, eos = "unesco")
#> [1] 1026.173
swRho(34, swTheta(34, 10, 100, 200, eos = "unesco"), 200, eos = "unesco")
#> [1] 1027.074
plotTS(as.ctd(c(30, 40), c(-2, 20), rep(0, 2)), eos = "unesco", grid = TRUE, col = "white")and in the Gibbs SeaWater system, we use eos="gws" and
must supply longitude and latitude arguments
to the sw*() calls and also to the as.ctd()
call.
Exercise 3. Draw a
-
diagram for the section data, using black symbols to the east of 30W and
gray symbols to the west, thus highlighting Mediterranean-derived
waters. Use handleFlags() (see using
data-quality flags) to discard questionable data, and use the
accessor function [[.
We will use the Gibbs SeaWater system, so as.ctd() needs
location information.
library(oce)
data(section)
s <- handleFlags(section, flags = list(c(1, 3:9)))
ctd <- as.ctd(s[["salinity"]], s[["temperature"]], s[["pressure"]],
longitude = s[["longitude"]], latitude = s[["latitude"]]
)
col <- ifelse(s[["longitude"]] > -30, "black", "gray")
plotTS(ctd, col = col, eos = "gsw")Exercise 4. Plot dynamic height and geostrophic
velocity across the Gulf Stream. (Hint: use the
swDynamicHeight function.)
(Try ?swDynamicHeight for hints on smoothing.)
library(oce)
data(section)
GS <- subset(section, 102 <= stationId & stationId <= 124)
dh <- swDynamicHeight(GS)
#> Warning in regularize.values(x, y, ties, missing(ties), na.rm = na.rm):
#> collapsing to unique 'x' values
par(mfrow = c(2, 1), mar = c(3, 3, 1, 1), mgp = c(2, 0.7, 0))
plot(dh$distance, dh$height, type = "l", xlab = "", ylab = "Dyn. Height [m]")
grid()
# 1e3 metres per kilometre
latMean <- mean(GS[["latitude"]])
f <- coriolis(latMean)
g <- gravity(latMean)
v <- diff(dh$height) / diff(dh$distance) * g / f / 1e3
plot(dh$distance[-1], v, type = "l", xlab = "Distance [km]", ylab = "Velocity [m/s]")
grid()
abline(h = 0, col = "red")References
Kelley, Dan E. Oceanographic Analysis with R. 1st ed. 2018. New York, NY: Springer New York : Imprint: Springer, 2018. https://doi.org/10.1007/978-1-4939-8844-0.
Kelley, Dan E., Clark Richards, and Chantelle Layton. “Oce: An R Package for Oceanographic Analysis.” Journal of Open Source Software 7, no. 71 (March 3, 2022): 3594. https://doi.org/10.21105/joss.03594.