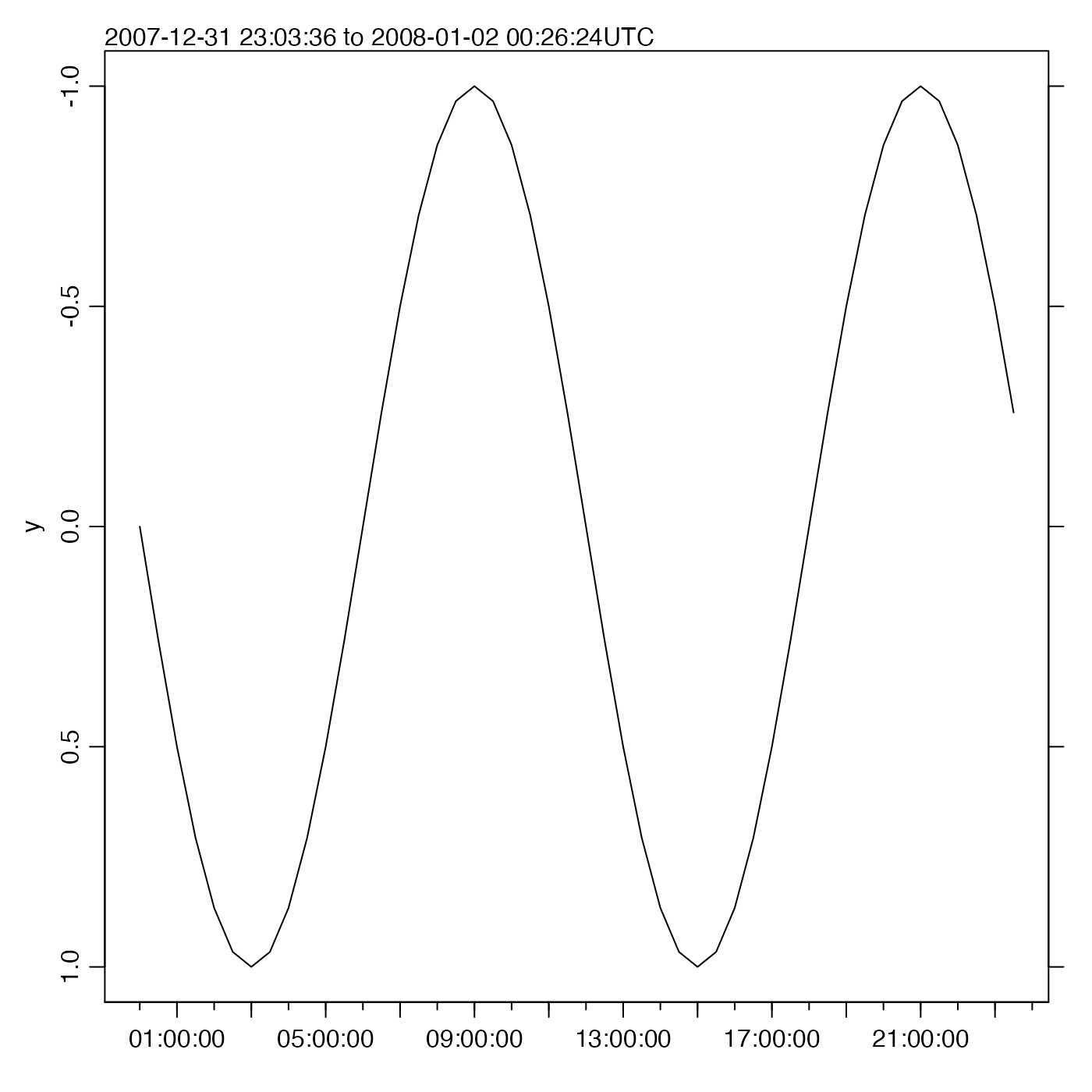
Plot a time-series, obeying the timezone and possibly drawing the range in the top-left margin.
Usage
oce.plot.ts(
x,
y,
type = "l",
xlim,
ylim,
log = "",
logStyle = "r",
flipy = FALSE,
xlab = "",
ylab,
drawTimeRange,
simplify = 2560,
fill = FALSE,
col = par("col"),
pch = par("pch"),
cex = par("cex"),
cex.axis = par("cex.axis"),
cex.lab = par("cex.lab"),
cex.main = par("cex.main"),
xaxs = par("xaxs"),
yaxs = par("yaxs"),
mgp = getOption("oceMgp"),
mar = c(mgp[1] + if (nchar(xlab) > 0) 1.5 else 1, mgp[1] + 1.5, mgp[2] + 1, mgp[2] +
3/4),
main = "",
despike = FALSE,
axes = TRUE,
tformat,
marginsAsImage = FALSE,
grid = FALSE,
grid.col = "lightgray",
grid.lty = "dotted",
grid.lwd = par("lwd"),
debug = getOption("oceDebug"),
...
)Arguments
- x
the times of observations. If this is not a POSIXt object, then an attempt is made to convert it to one using
as.POSIXct().- y
the observations.
- type
plot type,
"l"for lines,"p"for points.- xlim
optional limit for x axis. This has an additional effect, beyond that for conventional R functions: it effectively windows the data, so that autoscaling will yield limits for y that make sense within the window.
- ylim
optional limit for y axis.
- log
a character value that must be either empty (the default) for linear
yaxis, or"y"for logarithmicyaxis. (Unlikeplot.default()etc.,oce.plot.tsdoes not permit logarithmic time, orxaxis.)- logStyle
a character value that indicates how to draw the y axis, if
log="y". If it is"r"(the default) then the conventional R style is used, in which a logarithmic transform connects y values to position on the "page" of the plot device, so that tics will be nonlinearly spaced, but not organized by integral powers of 10. However, if it is"decade", then the style will be that used in the scientific literature, in which large tick marks are used for integral powers of 10, with smaller tick marks at integral multiples of those powers, and with labels that use exponential format for values above 100 or below 0.01. The value oflogStyleis passed tooceAxis(), which draws the axis.- flipy
Logical, with
TRUEindicating that the graph should have the y axis reversed, i.e. with smaller values at the bottom of the page.- xlab
name for x axis; defaults to
"".- ylab
name for y axis; defaults to the plotted item.
- drawTimeRange
an optional indication of whether/how to draw a time range, in the top-left margin of the plot; see
oce.axis.POSIXct()for details.- simplify
an integer value that indicates whether to speed up
type="l"plots by replacing the data with minimum and maximum values within a subsampled time mesh. This can speed up plots of large datasets (e.g. by factor 20 for 10^7 points), sometimes with minor changes in appearance. This procedure is skipped ifsimplifyisNAor if the number of visible data points is less than 5 timessimplify. Otherwise,oce.plot.tscreatessimplifyintervals ranging across the visible time range. Intervals with under 2 finiteydata are ignored. In the rest,yvalues are replaced with their range, andxvalues are replaced with the repeated midpoint time. Thus, each retained sub-interval has exactly 2 data points. A warning is printed if this replacement is done. The default value ofsimplifymeans that cases with under 2560 visible points are plotted conventionally.- fill
boolean, set
TRUEto fill the curve to zero (which it does incorrectly if there are missing values iny).- col
The colours for points (if
type=="p") or lines (iftype=="l"). For thetype="p"case, if there are fewercolvalues than there arexvalues, then thecolvalues are recycled in the standard fashion. For thetype="l"case, the line is plotted in the first colour specified.- pch
character code, used if
type=="p". If there are fewerpchvalues than there arexvalues, then thepchvalues are recycled in the standard fashion. Seepoints()for the possible values forpch.- cex
numeric character expansion factor for points on plots, ignored unless
typeis"p". This may be a single number, applied to all points, or a vector of numbers to be applied to the points in sequence. If there are fewerpchvalues than there arexvalues, then thepchvalues are recycled in the standard fashion. Seepar()for more oncex.- cex.axis, cex.lab, cex.main
numeric character expansion factors for axis numbers, axis names and plot titles; see
par().- xaxs
control x axis ending; see
par("xaxs").- yaxs
control y axis ending; see
par("yaxs").- mgp
3-element numerical vector to use for
par(mgp), and also forpar(mar), computed from this. The default is tighter than the R default, in order to use more space for the data and less for the axes.- mar
value to be used with
par("mar")to set margins. The default value uses significantly tighter margins than is the norm in R, which gives more space for the data. However, in doing this, the existingpar("mar")value is ignored, which contradicts values that may have been set by a previous call todrawPalette(). To get plot with a palette, first calldrawPalette(), then calloce.plot.tswithmar=par("mar").- main
title of plot.
- despike
boolean flag that can turn on despiking with
despike().- axes
boolean, set to
TRUEto get axes plotted- tformat
optional format for labels on the time axis
- marginsAsImage
logical value indicating whether to set the right-hand margin to the width normally taken by an image drawn with
imagep().- grid
logical value indicating whether to draw a grid on the plot. For time axes labelled in months, this will differ from the grid that would be drawn by calling
grid()after the plot was completed, becauseoce.plot.ts()draws months with appropriate relative lengths, whereas R time axes consider months to be of equal length.- grid.col
color of grid
- grid.lty
line type of grid
- grid.lwd
line width of grid
- debug
a flag that turns on debugging. Set to 1 to get a moderate amount of debugging information, or to 2 to get more.
- ...
graphical parameters passed down to
plot().
Value
A list is silently returned, containing xat and yat,
values that can be used by oce.grid() to add a grid to the plot.
Details
Depending on the version of R, the standard plot() and
plot.ts() routines will not obey the time zone of the data.
This routine gets around that problem. It can also plot the time range in
the top-left margin, if desired; this string includes the timezone, to
remove any possible confusion.
The time axis is drawn with oce.axis.POSIXct().
Examples
library(oce)
t0 <- as.POSIXct("2008-01-01", tz = "UTC")
t <- seq(t0, length.out = 48, by = "30 min")
y <- sin(as.numeric(t - t0) * 2 * pi / (12 * 3600))
oce.plot.ts(t, y, type = "l", xaxs = "i")
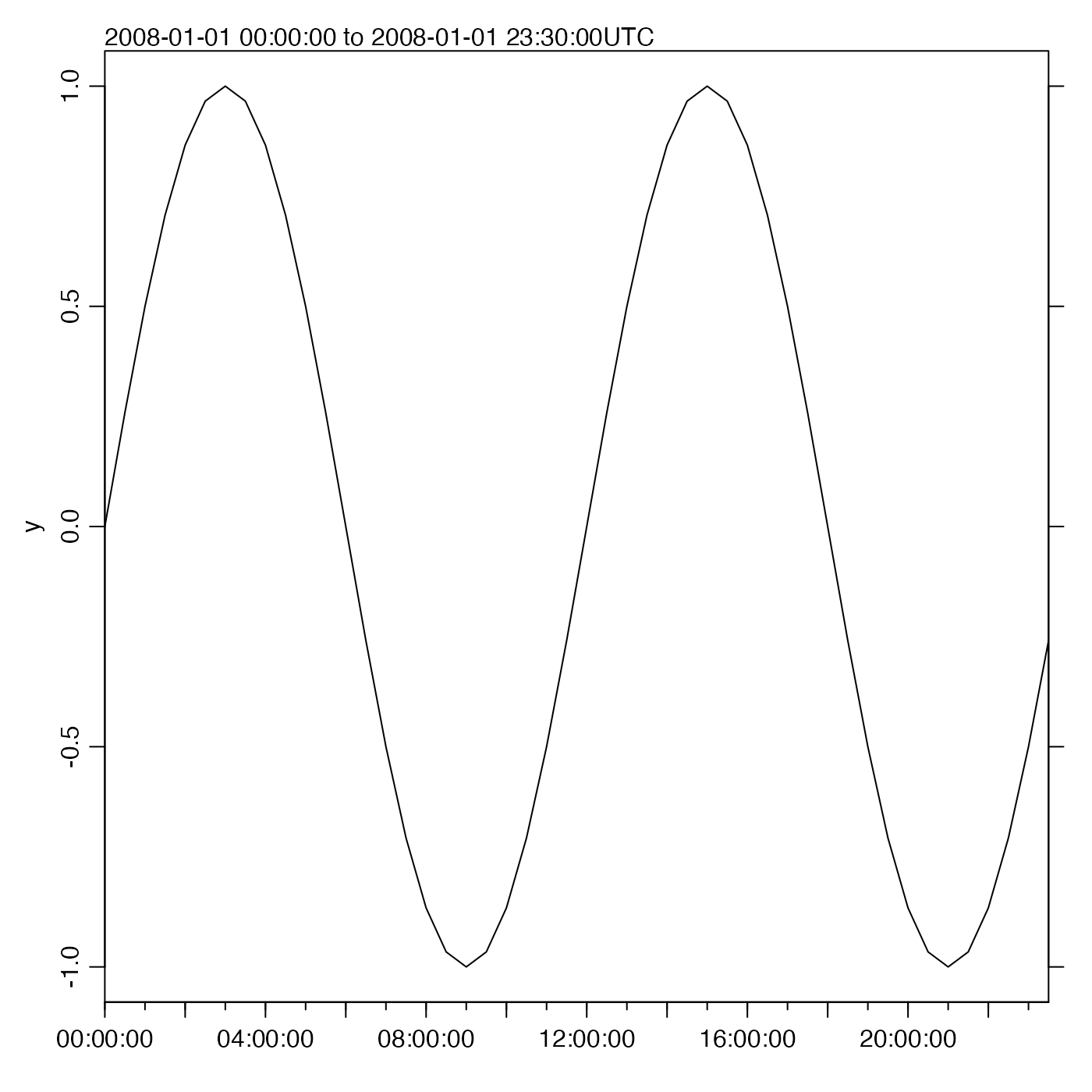 # Show how col, pch and cex get recycled
oce.plot.ts(t, y,
type = "p", xaxs = "i",
col = 1:3, pch = c(rep(1, 6), rep(20, 6)), cex = sqrt(1:6)
)
# Show how col, pch and cex get recycled
oce.plot.ts(t, y,
type = "p", xaxs = "i",
col = 1:3, pch = c(rep(1, 6), rep(20, 6)), cex = sqrt(1:6)
)
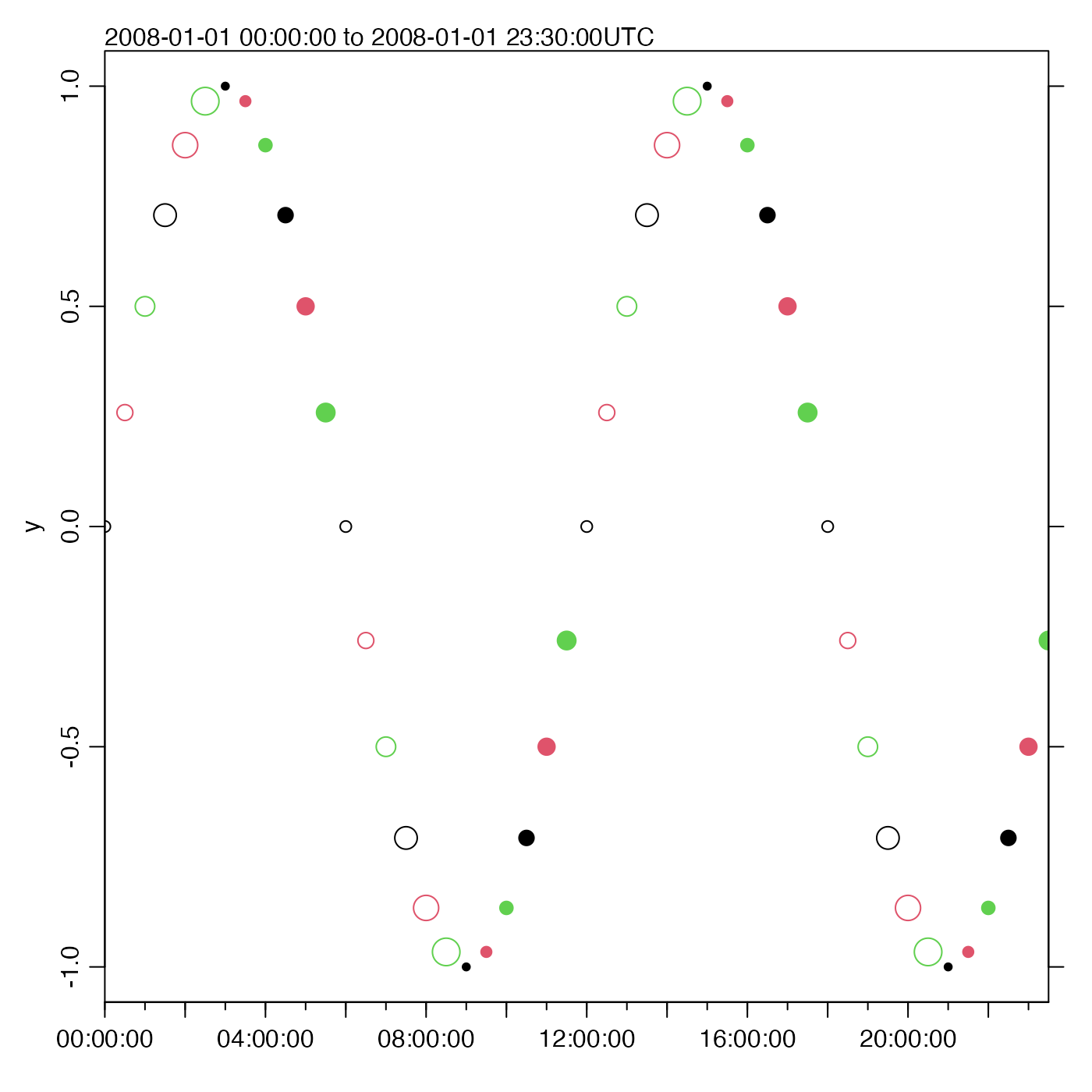 # Trimming x; note the narrowing of the y view
oce.plot.ts(t, y, type = "p", xlim = c(t[6], t[12]))
# Trimming x; note the narrowing of the y view
oce.plot.ts(t, y, type = "p", xlim = c(t[6], t[12]))
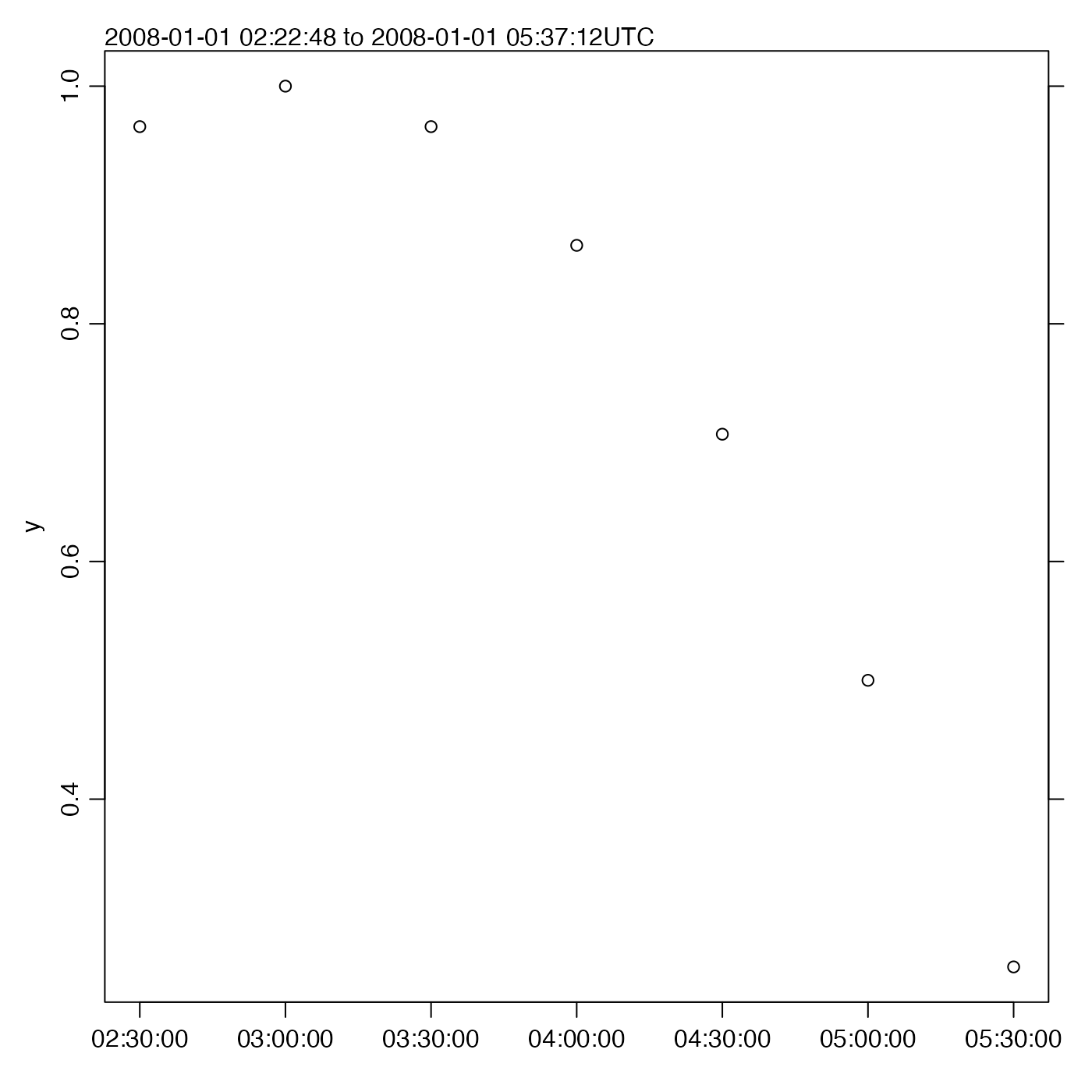 # Flip the y axis
oce.plot.ts(t, y, flipy = TRUE)
# Flip the y axis
oce.plot.ts(t, y, flipy = TRUE)