This works by calling ncdf2oce() and then using class() on
the result to make it be of subclass "adv". This is intended
to work with NetCDF files created with adv2ncdf(), which embeds
sufficient information in the file to permit ncdf2adv() to
reconstruct the original adv object. See the documentation
for adv2ncdf() to learn more about what it stores, and therefore
what ncdf2adv() attempts to read.
ncdf2adv(ncfile = NULL, varTable = NULL, debug = 0)Arguments
- ncfile
character value naming the input file.
- varTable
character value indicating the variable-naming scheme to be used, which is passed to
read.varTable()to set up variable names, units, etc.- debug
integer, 0 (the default) for quiet action apart from messages and warnings, or any larger value to see more output that describes the processing steps.
Value
ncdf2adv() returns an adv object.
Examples
library(ocencdf)
# Example with an adv file from oce package
data(adv, package = "oce")
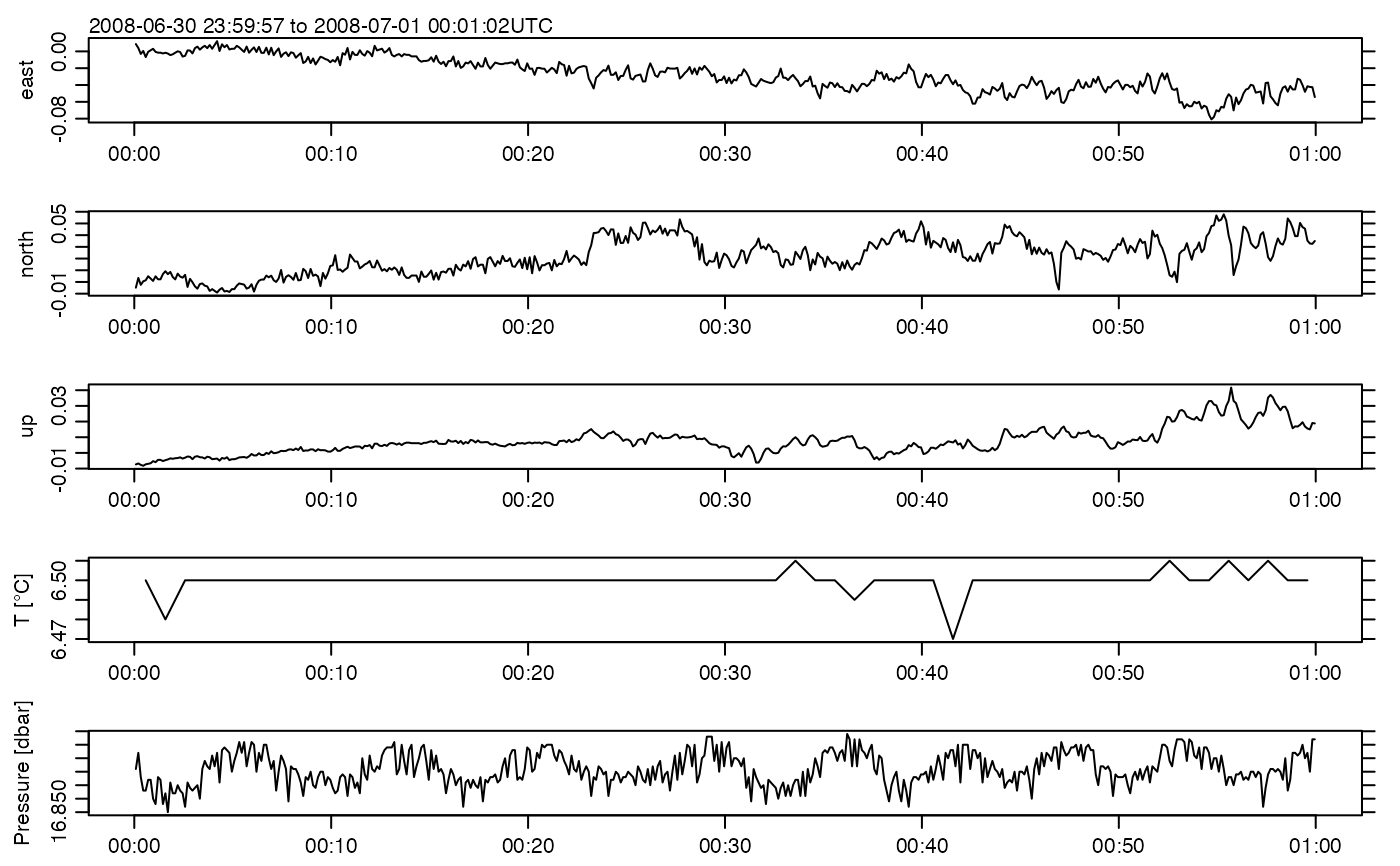
summary(adv)
#> ADV Summary
#> -----------
#>
#> * Instrument: vector, serial number ``(serial number redacted)``
#> * Filename: `(file name redacted)`
#> * Location: 47.87943 N , -69.72533 E
#> * Time: 2008-07-01 00:00:00 to 2008-07-01 00:00:59 (480 samples, mean increment 0.1250001 s)
#> * Data Overview
#>
#> Min. Mean Max. Dim. NAs
#> v [m/s] -0.080871 0.00069514 0.057789 480x3 0
#> a NA NA NA 480x3 0
#> q NA NA NA 480x3 0
#> time 2008-07-01 00:00:00 2008-07-01 00:00:30 2008-07-01 00:00:59 480 0
#> pressure [dbar] 16.85 16.866 16.879 480 0
#> timeBurst NA NA NA 480 480
#> recordsBurst NA NA NA 480 480
#> voltageSlow 9.7 9.71 9.8 60 0
#> timeSlow 1214870401 1214870430 1214870460 60 0
#> headingSlow [°] -23.39 -23.39 -23.39 60 0
#> pitchSlow [°] 0.4 0.5 0.6 60 0
#> rollSlow [°] -6.2 -6.145 -6.1 60 0
#> temperatureSlow [°C] 6.47 6.4997 6.51 60 0
#>
#> * Processing Log
#>
#> - 2015-12-23 17:53:39 UTC: `read.oce(file = "/data/archive/sleiwex/2008/moorings/m05/adv/nortek_1943/raw/adv_nortek_1943.vec", from = as.POSIXct("2008-06-25 00:00:00", tz = "UTC"), to = as.POSIXct("2008-07-06 00:00:00", tz = "UTC"), latitude = 47.87943, longitude = -69.72533)`
#> - 2015-12-23 17:53:54 UTC: `retime(x = m05VectorBeam, a = 0.58, b = 6.3892e-07, t0 = as.POSIXct("2008-07-01 00:00:00", tz = "UTC"))`
#> - 2015-12-23 17:53:55 UTC: `subset(x, subset=as.POSIXct("2008-06-25 13:00:00", tz = "UTC") <= time & time <= as.POSIXct("2008-07-03 00:50:00", tz = "UTC"))`
#> - 2015-12-23 17:53:55 UTC: `oceEdit(x = m05VectorBeam, item = "transformationMatrix", value = rbind(c(11033, -5803, -5238), c(347, -9622, 9338), c(-1418, -1476, -1333))/4096, reason = "Nortek email 2011-02-14", person = "DEK")`
#> - 2015-12-23 17:53:55 UTC: `use aquadoppHR heading; despike own pitch and roll`
#> - 2015-12-23 17:54:11 UTC: `beamToXyzAdv(x = x)`
#> - 2015-12-23 17:54:34 UTC: `xyzToEnu(x, declination=-18.099, horizontalCase=TRUE, sensorOrientiation=upward, debug=0)`
plot(adv)
 # Transfer to NetCDF and back to see if results make sense
# Use a temporary nc file to let package pass CRAN checks.
ncfile <- tempfile(pattern = "adv", fileext = ".nc")
oce2ncdf(adv, ncfile = ncfile)
ADV <- ncdf2adv(ncfile)
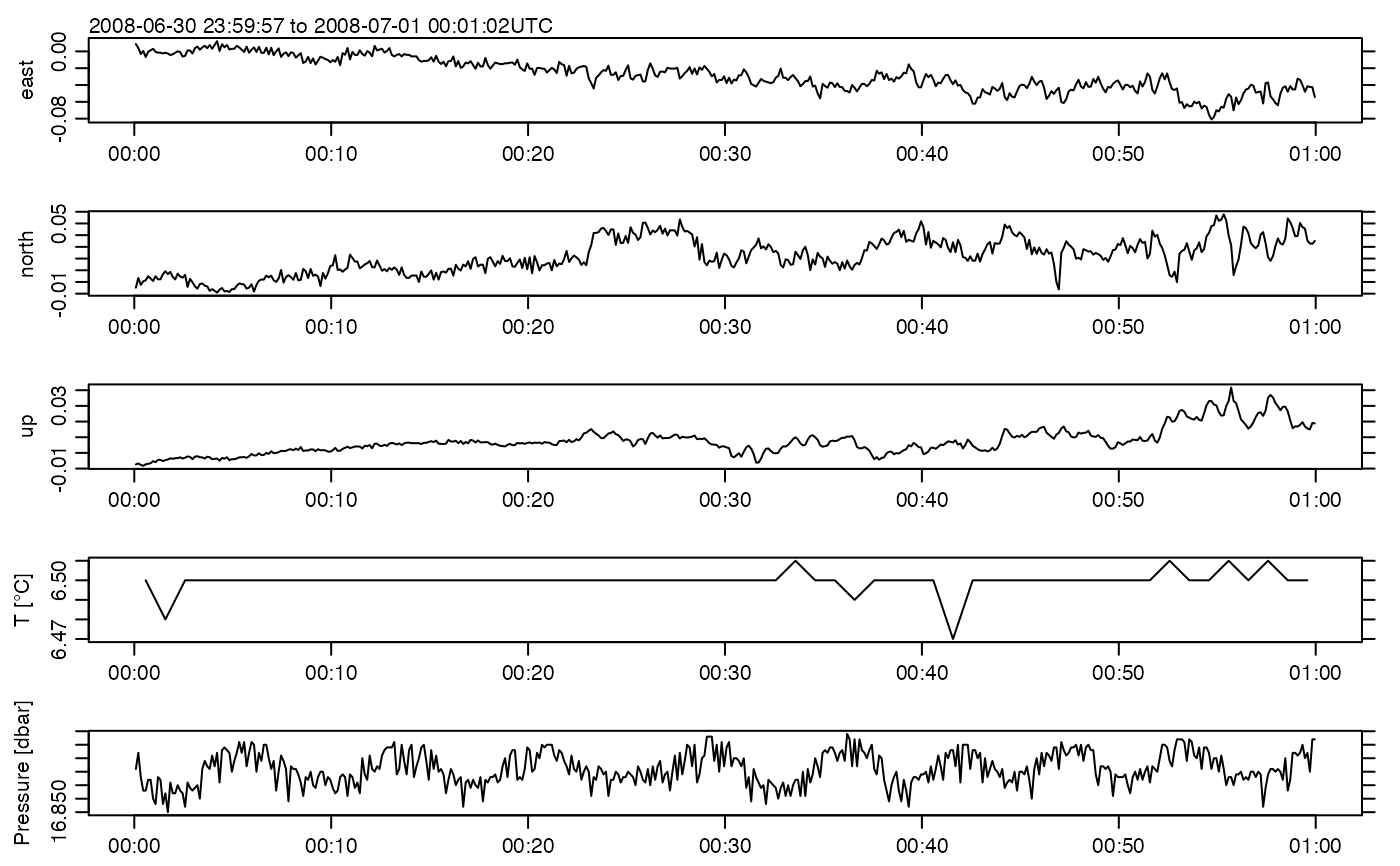
summary(ADV)
#> ADV Summary
#> -----------
#>
#> * Instrument: vector, serial number ``(serial number redacted)``
#> * Filename: `(file name redacted)`
#> * Location: 47.87943 N , -69.72533 E
#> * Time: 2008-07-01 00:00:00 to 2008-07-01 00:00:59 (480 samples, mean increment 0.1250001 s)
#> * Data Overview
#>
#> Min. Mean Max. Dim. NAs
#> v [m/s] -0.080871 0.00069514 0.057789 480x3 0
#> a NA NA NA 480x3 0
#> q NA NA NA 480x3 0
#> time 2008-07-01 00:00:00 2008-07-01 00:00:30 2008-07-01 00:00:59 480 0
#> pressure [dbar] 16.85 16.866 16.879 480 0
#> timeBurst NA NA NA 480 480
#> recordsBurst NA NA NA 480 480
#> voltageSlow 9.7 9.71 9.8 60 0
#> timeSlow 1214870401 1214870430 1214870460 60 0
#> headingSlow [°] -23.39 -23.39 -23.39 60 0
#> pitchSlow [°] 0.4 0.5 0.6 60 0
#> rollSlow [°] -6.2 -6.145 -6.1 60 0
#> temperatureSlow [°C] 6.47 6.4997 6.51 60 0
#>
#> * Processing Log
#>
#> - 2024-02-22 00:13:57 UTC: `Create oce object`
plot(ADV)
# Transfer to NetCDF and back to see if results make sense
# Use a temporary nc file to let package pass CRAN checks.
ncfile <- tempfile(pattern = "adv", fileext = ".nc")
oce2ncdf(adv, ncfile = ncfile)
ADV <- ncdf2adv(ncfile)
summary(ADV)
#> ADV Summary
#> -----------
#>
#> * Instrument: vector, serial number ``(serial number redacted)``
#> * Filename: `(file name redacted)`
#> * Location: 47.87943 N , -69.72533 E
#> * Time: 2008-07-01 00:00:00 to 2008-07-01 00:00:59 (480 samples, mean increment 0.1250001 s)
#> * Data Overview
#>
#> Min. Mean Max. Dim. NAs
#> v [m/s] -0.080871 0.00069514 0.057789 480x3 0
#> a NA NA NA 480x3 0
#> q NA NA NA 480x3 0
#> time 2008-07-01 00:00:00 2008-07-01 00:00:30 2008-07-01 00:00:59 480 0
#> pressure [dbar] 16.85 16.866 16.879 480 0
#> timeBurst NA NA NA 480 480
#> recordsBurst NA NA NA 480 480
#> voltageSlow 9.7 9.71 9.8 60 0
#> timeSlow 1214870401 1214870430 1214870460 60 0
#> headingSlow [°] -23.39 -23.39 -23.39 60 0
#> pitchSlow [°] 0.4 0.5 0.6 60 0
#> rollSlow [°] -6.2 -6.145 -6.1 60 0
#> temperatureSlow [°C] 6.47 6.4997 6.51 60 0
#>
#> * Processing Log
#>
#> - 2024-02-22 00:13:57 UTC: `Create oce object`
plot(ADV)
 file.remove(ncfile)
#> [1] TRUE
file.remove(ncfile)
#> [1] TRUE