Given an adv object created by the oce package, this function
creates a NetCDF file that can later by read by ncdf2adv() to approximately
reproduce the original contents.
adv2ncdf(x, varTable = NULL, ncfile = NULL, force_v4 = TRUE, debug = 0)Arguments
- x
an oce object of class
adv, as created by e.g.oce::read.adv().- varTable
character value indicating the variable-naming scheme to be used, which is passed to
read.varTable()to set up variable names, units, etc.- ncfile
character value naming the output file. Use NULL for a file name to be created automatically (e.g.
ctd.ncfor a CTD object).- force_v4
logical value which controls the NetCDF file version during the nc_create step. The default here is TRUE, whereas the ncdf4-package defaults to FALSE (ensuring that the NetCDF file is compatible with NetCDF v3). Some features, including large data sizes, may require v4.
- debug
integer, 0 (the default) for quiet action apart from messages and warnings, or any larger value to see more output that describes the processing steps.
Details
Note that adv2ncdf() defaults varTable to "adv".
The entire contents of the metadata slot are saved in the global attribute named
"metadata", in a JSON format. The JSON material is developed with
metadata2json(), which yields a value that can be decoded with
json2metadata().
See also
Other things related to adv data:
ncdf2adv()
Examples
library(ocencdf)
# Example with an adv file from oce package
data(adv, package = "oce")
summary(adv)
#> ADV Summary
#> -----------
#>
#> * Instrument: vector, serial number ``(serial number redacted)``
#> * Filename: `(file name redacted)`
#> * Location: 47.87943 N , -69.72533 E
#> * Time: 2008-07-01 00:00:00 to 2008-07-01 00:00:59 (480 samples, mean increment 0.1250001 s)
#> * Data Overview
#>
#> Min. Mean Max. Dim. NAs
#> v [m/s] -0.080871 0.00069514 0.057789 480x3 0
#> a NA NA NA 480x3 0
#> q NA NA NA 480x3 0
#> time 2008-07-01 00:00:00 2008-07-01 00:00:30 2008-07-01 00:00:59 480 0
#> pressure [dbar] 16.85 16.866 16.879 480 0
#> timeBurst NA NA NA 480 480
#> recordsBurst NA NA NA 480 480
#> voltageSlow 9.7 9.71 9.8 60 0
#> timeSlow 1214870401 1214870430 1214870460 60 0
#> headingSlow [°] -23.39 -23.39 -23.39 60 0
#> pitchSlow [°] 0.4 0.5 0.6 60 0
#> rollSlow [°] -6.2 -6.145 -6.1 60 0
#> temperatureSlow [°C] 6.47 6.4997 6.51 60 0
#>
#> * Processing Log
#>
#> - 2015-12-23 17:53:39 UTC: `read.oce(file = "/data/archive/sleiwex/2008/moorings/m05/adv/nortek_1943/raw/adv_nortek_1943.vec", from = as.POSIXct("2008-06-25 00:00:00", tz = "UTC"), to = as.POSIXct("2008-07-06 00:00:00", tz = "UTC"), latitude = 47.87943, longitude = -69.72533)`
#> - 2015-12-23 17:53:54 UTC: `retime(x = m05VectorBeam, a = 0.58, b = 6.3892e-07, t0 = as.POSIXct("2008-07-01 00:00:00", tz = "UTC"))`
#> - 2015-12-23 17:53:55 UTC: `subset(x, subset=as.POSIXct("2008-06-25 13:00:00", tz = "UTC") <= time & time <= as.POSIXct("2008-07-03 00:50:00", tz = "UTC"))`
#> - 2015-12-23 17:53:55 UTC: `oceEdit(x = m05VectorBeam, item = "transformationMatrix", value = rbind(c(11033, -5803, -5238), c(347, -9622, 9338), c(-1418, -1476, -1333))/4096, reason = "Nortek email 2011-02-14", person = "DEK")`
#> - 2015-12-23 17:53:55 UTC: `use aquadoppHR heading; despike own pitch and roll`
#> - 2015-12-23 17:54:11 UTC: `beamToXyzAdv(x = x)`
#> - 2015-12-23 17:54:34 UTC: `xyzToEnu(x, declination=-18.099, horizontalCase=TRUE, sensorOrientiation=upward, debug=0)`
plot(adv)
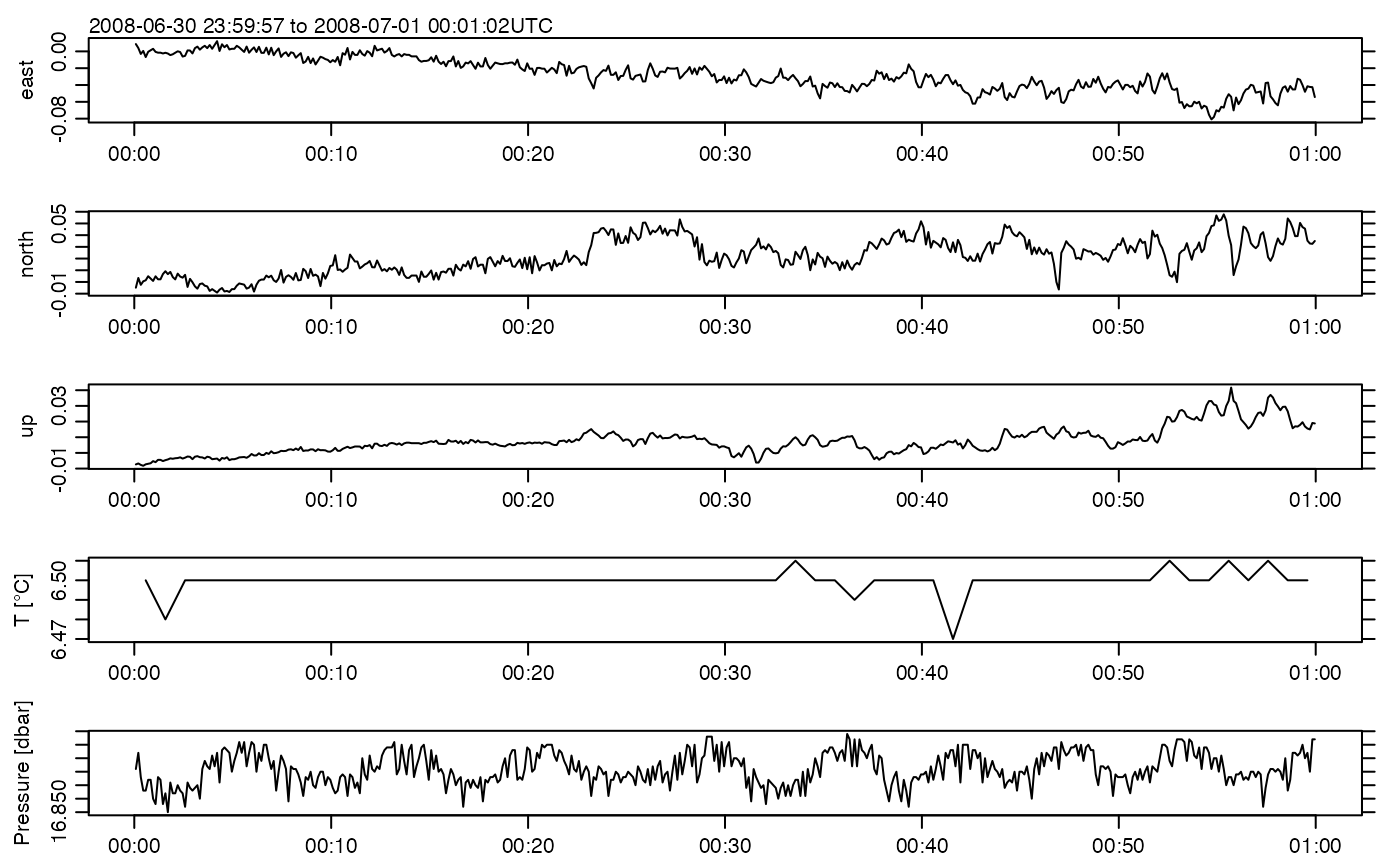 # Transfer to NetCDF and back to see if results make sense
# Use a temporary nc file to let package pass CRAN checks.
ncfile <- tempfile(pattern = "adv", fileext = ".nc")
oce2ncdf(adv, ncfile = ncfile)
ADV <- ncdf2adv(ncfile)
summary(ADV)
#> ADV Summary
#> -----------
#>
#> * Instrument: vector, serial number ``(serial number redacted)``
#> * Filename: `(file name redacted)`
#> * Location: 47.87943 N , -69.72533 E
#> * Time: 2008-07-01 00:00:00 to 2008-07-01 00:00:59 (480 samples, mean increment 0.1250001 s)
#> * Data Overview
#>
#> Min. Mean Max. Dim. NAs
#> v [m/s] -0.080871 0.00069514 0.057789 480x3 0
#> a NA NA NA 480x3 0
#> q NA NA NA 480x3 0
#> time 2008-07-01 00:00:00 2008-07-01 00:00:30 2008-07-01 00:00:59 480 0
#> pressure [dbar] 16.85 16.866 16.879 480 0
#> timeBurst NA NA NA 480 480
#> recordsBurst NA NA NA 480 480
#> voltageSlow 9.7 9.71 9.8 60 0
#> timeSlow 1214870401 1214870430 1214870460 60 0
#> headingSlow [°] -23.39 -23.39 -23.39 60 0
#> pitchSlow [°] 0.4 0.5 0.6 60 0
#> rollSlow [°] -6.2 -6.145 -6.1 60 0
#> temperatureSlow [°C] 6.47 6.4997 6.51 60 0
#>
#> * Processing Log
#>
#> - 2024-02-22 00:13:50 UTC: `Create oce object`
plot(ADV)
# Transfer to NetCDF and back to see if results make sense
# Use a temporary nc file to let package pass CRAN checks.
ncfile <- tempfile(pattern = "adv", fileext = ".nc")
oce2ncdf(adv, ncfile = ncfile)
ADV <- ncdf2adv(ncfile)
summary(ADV)
#> ADV Summary
#> -----------
#>
#> * Instrument: vector, serial number ``(serial number redacted)``
#> * Filename: `(file name redacted)`
#> * Location: 47.87943 N , -69.72533 E
#> * Time: 2008-07-01 00:00:00 to 2008-07-01 00:00:59 (480 samples, mean increment 0.1250001 s)
#> * Data Overview
#>
#> Min. Mean Max. Dim. NAs
#> v [m/s] -0.080871 0.00069514 0.057789 480x3 0
#> a NA NA NA 480x3 0
#> q NA NA NA 480x3 0
#> time 2008-07-01 00:00:00 2008-07-01 00:00:30 2008-07-01 00:00:59 480 0
#> pressure [dbar] 16.85 16.866 16.879 480 0
#> timeBurst NA NA NA 480 480
#> recordsBurst NA NA NA 480 480
#> voltageSlow 9.7 9.71 9.8 60 0
#> timeSlow 1214870401 1214870430 1214870460 60 0
#> headingSlow [°] -23.39 -23.39 -23.39 60 0
#> pitchSlow [°] 0.4 0.5 0.6 60 0
#> rollSlow [°] -6.2 -6.145 -6.1 60 0
#> temperatureSlow [°C] 6.47 6.4997 6.51 60 0
#>
#> * Processing Log
#>
#> - 2024-02-22 00:13:50 UTC: `Create oce object`
plot(ADV)
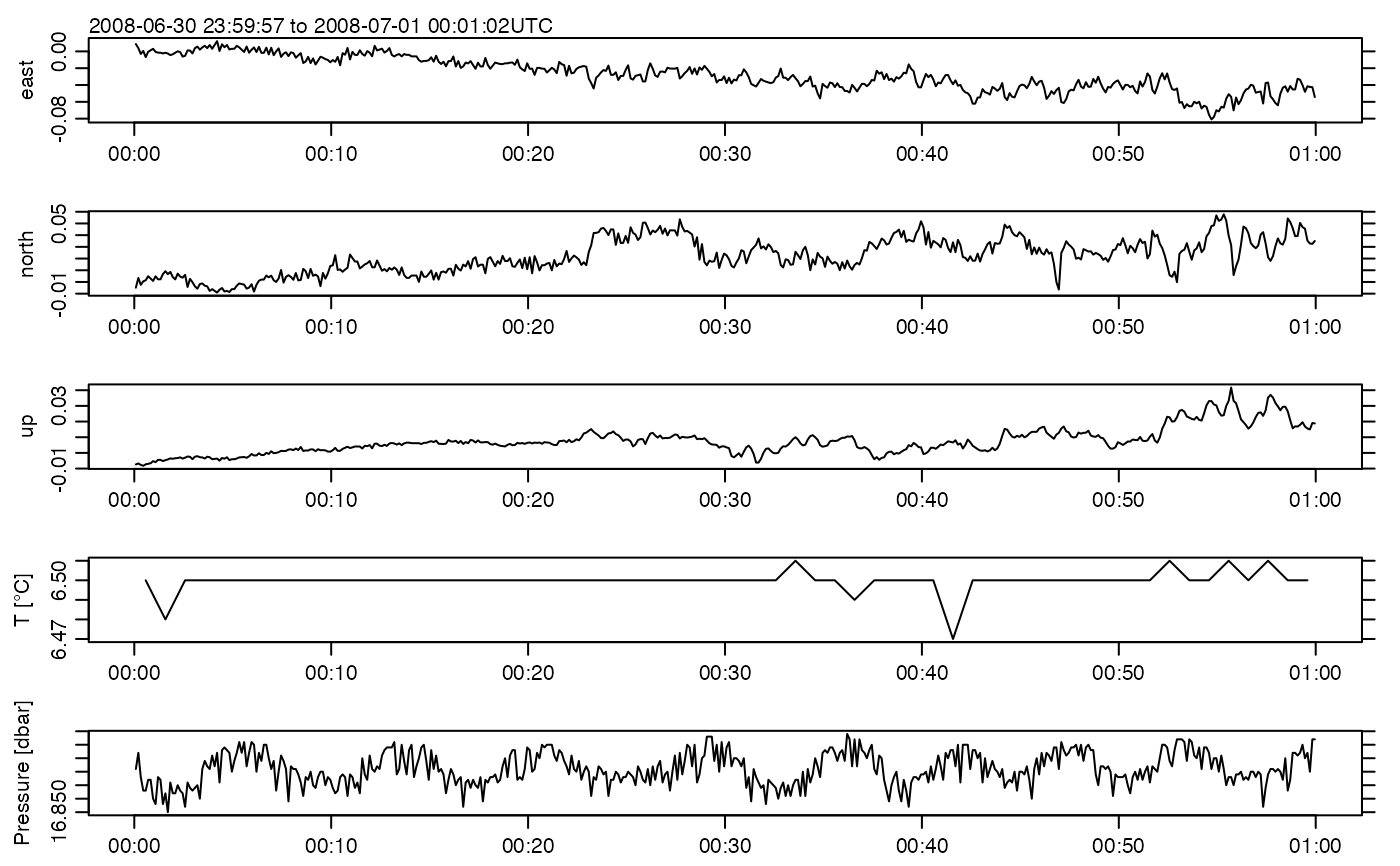 file.remove(ncfile)
#> [1] TRUE
file.remove(ncfile)
#> [1] TRUE