Introduction
I was doing some thinking about how best to work with rotating spectra in R,
and wanted to drop all the way down to using fft() instead of spectrum().
For a while, I was confused about some of the results, because the lengths of
spectra created with fft were not what I expected. Then I saw that the
problem was that I was using the default value of fast in the spectrum()
function. The content of this post illustrates this.
Results and discussion
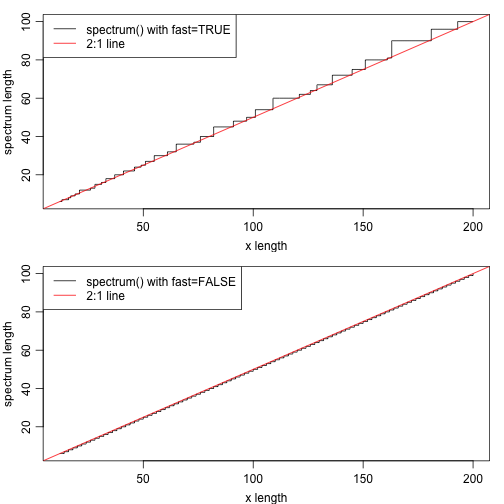
By default, the R spectrum() function sets the argument fast=TRUE. This
causes R to increase the speed of processing, but it also leads to spectrum
lengths that can be surprising.
The following illustrates how the length of spectra computed with spectrum
vary from the expected value of floor(length(data)/2).
1
2
3
4
5
6
7
8
9
10
11
12
13
## below shows that spectrum() does some tricky things
par(mfrow=c(2, 1), mar=c(3, 3, 1, 1), mgp=c(2, 0.7, 0))
set.seed(123)
x <- 12:200
## fast=TRUE is the default
speclenT <- unlist(lapply(x, function(n) length(spec.pgram(rnorm(n),plot=FALSE,fast=TRUE)$freq)))
plot(x, speclenT, type="s", xlab="x length", ylab="spectrum length")
legend("topleft", lwd=par("lwd"), col=1:2, legend=c("spectrum() with fast=TRUE", "2:1 line"))
abline(0, 1/2, col=2)
speclenF <- unlist(lapply(x, function(n) length(spec.pgram(rnorm(n),plot=FALSE,fast=FALSE)$freq)))
plot(x, speclenF, type="s", xlab="x length", ylab="spectrum length")
legend("topleft", lwd=par("lwd"), col=1:2, legend=c("spectrum() with fast=FALSE", "2:1 line"))
abline(0, 1/2, col=2)
1
print(head(speclenT))
## [1] 6 7 7 7 8 91
print(head(speclenF))
## [1] 6 6 7 7 8 81
all.equal(speclenF, floor(x/2))
## [1] TRUEConclusion
If the results of spectrum and fft are to be put on an equal footing, e.g. for numerical
comparisons, then it makes sense to call the former as e.g. spectrum(..., fast=FALSE).
Reference and resources
- Jekyll source code for this blog entry: 2017-12-21-spectrum-length.Rmd